Protein AssociatioN Energy Landscape (PANEL)
PANEL scripts for download
Cite:
Rajagopal, N.; Nangia, S. Obtaining Protein Association Energy Landscape for Integral Membrane Proteins. Journal of Chemical Theory and Computation 2019.
DOI:10.1021/acs.jctc.9b00626
Background
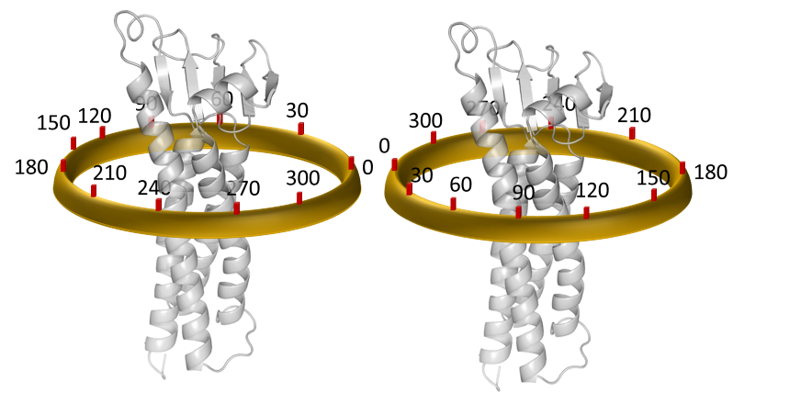
Protein AssociatioN Energy Landscape (PANEL) provides an extensive and converged data set for all possible conformations of integral membrane protein associations using a combination of stochastic sampling and equilibration simulations. The PANEL method samples the rotational space around each interacting proteins to obtain the comprehensive interaction energy landscape.
Gram Negative Bacteria
Force field parameters
- Simulating gram-negative bacterial outer membrane: A coarse grain model, H. Ma, F. J. Irudayanathan, W. Jiang, and S. Nangia, Journal of Physical Chemistry B, 119, 14668–14682 (2015).
- Modeling diversity in structures of bacterial outer membrane lipids, H. Ma, D. D. Cummins, N. B. Edelstein, J. Gomez, A. Khan, M. D. Llewellyn, T. Picudella, S. R. Willsey, and S. Nangia, Journal of Chemical Theory and Computation, 13, 811–824 (2017).
– A coarse grained force field for outer membrane that is computationally affordable for simulating dynamical process over physiologically relevant time scales.
