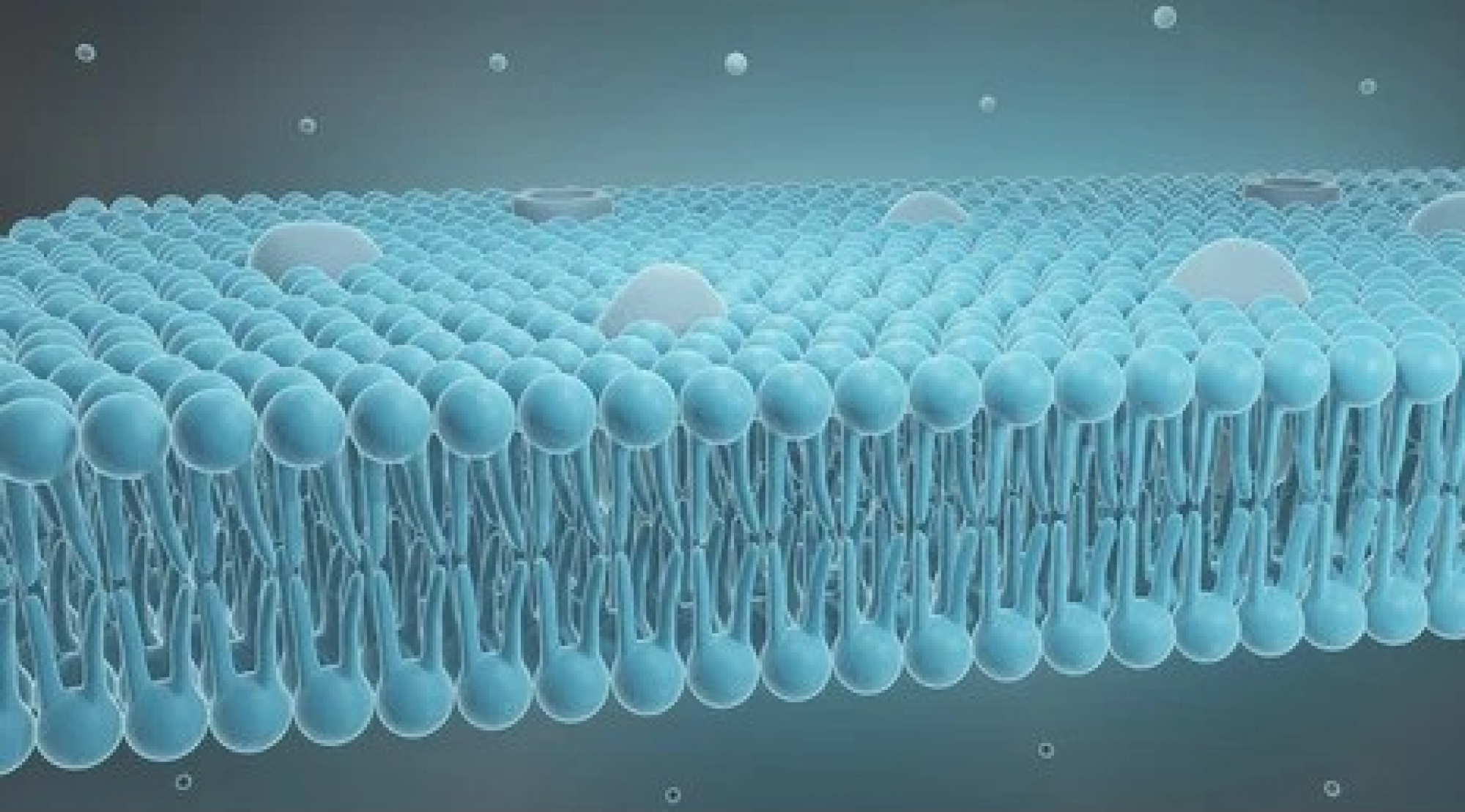
The blood-brain barrier serves the critical role of allowing only certain types of molecules to enter the brain from the bloodstream. This important capability protects the brain from exposure to harmful chemical compounds. However, it also prevents certain drugs from entering the brain that treats brain conditions or diseases such as Alzheimer’s. Since the segment of the US population older than 65 is expected to increase by 50% by 2030, and the cost of care for these kinds of brain diseases is on the order of billions of dollars per year, finding new ways to help drugs cross the blood-brain barrier is urgently needed. However, understanding how therapeutic drug molecules move or do not move across the barrier into the brain has remained elusive.
In the past few years, we have developed a molecular model of the tight junction architecture of the tight junctions at the blood-brain interface. This model is now used to develop new strategies to deliver drug molecules to the brain.
Key Publications:
Computational Nanoscopy of Tight Junctions at the Blood–Brain Barrier Interface, N. Rajagopal, F. J. Irudayanathan, and S. Nangia, International Journal of Molecular Science, 20, 5583 (2019).
Obtaining Protein Association Energy Landscape for Integral Membrane Proteins, N. Rajagopal and S. Nangia, Journal of Chemical Theory and Computation, 15, 11, 6444-6455 (2019).
There is a growing urgency to prevent the misuse and overuse of antimicrobials to thwart the development of drug-resistant bacteria. Alternative strategies such as antimicrobial peptides, nanoparticles, and development of new materials that prevent bacterial colonization are needed to address the challenge.
Our group is working on various aspects of the antibiotic resistance challenge by developing models for the outer membranes of Gram-negative bacteria, membrane-penetrating ionic liquids, and on-demand antimicrobial microgels.
Key publications:
The influence of water on choline-based ionic liquids, E. E. L. Tanner, K. M. Piston, H. Ma, K. N. Ibsen, S. Nangia, S. Mitragotri, ACS Biomaterial Science and Engineering 5, 3645–3653 (2019).
Mechanism of Antibacterial Activity of Choline-Based Ionic Liquids (CAGE), Kelly N. Ibsen, H. Ma, A. Banerjee, E. E. L. Tanner, S. Nangia, and S. Mitragotri, ACS Biomaterials Science and Engineering 4, 2370–2379 (2018).
Modeling diversity in structures of bacterial outer membrane lipids, H. Ma, D. D. Cummins, N. B. Edelstein, J. Gomez, A. Khan, M. D. Llewellyn, T. Picudella, S. R. Willsey, and S. Nangia, Journal of Chemical Theory and Computation, 13, 811–824 (2017).
Elastin-like proteins (ELPs) are biopolymers with amino acid pentad of the general form (VPGXG)n, or Val-Pro-Gly-X-Gly, where “X” denotes a variable amino acid, and “n” denotes the number of monomeric units that comprise the polymer. The chemical charge and amphiphilicity of X impact the properties of the ELP, including the transition temperature (Tt). Fatty acid modification of the ELPs alters the hydrophilic/hydrophobic balance to create thermally responsive elastin-like polypeptides.
We model the fatty acid modified ELPs using advanced molecular dynamics using all-atom and coarse grain molecular dynamics techniques. This work is in collaboration with Mozhdehi Lab (Syracuse University).
Key Publications:
Adaptive Recombinant Nanoworms from Genetically Encodable Star Amphiphiles, MS Hossain, J Ji, CJ Lynch, M Guzman, S Nangia, D Mozhdehi, Biomacromolecules 23, 863-876 (2021).
Non-canonical lipoproteins with programmable assembly and architecture, MS Hossain, C Maller, Y Dai, S Nangia, D Mozhdehi, Chemical Communications 56 (71), 10281-10284 (2020).
Palmitoylation, a reversible posttranslational modification of a cysteine residue to palmitic acid (16:0) via a thioester bond. This modification has the unique ability to regulate protein dynamics and biological function. In membrane proteins, palmitoylation alters specific protein-protein and protein-lipid interactions, leading to changes in protein function. Palmitoylation affects receptor signaling, modified trafficking, reduced transporter activity, and ion-channel regulation, however, the biophysical effects are not well-understood. Our goal to use molecular modeling methods to characterize the impact of palmitoylation on protein’s structure, dynamics, and function.
Key Publications:
Palmitoylation of claudin-5 proteins Influences their lipid domain affinity and tight junction assembly at the blood-brain barrier interface, N. Rajagopal, F. J. Irudayanathan, and S. Nangia, Journal of Physical Chemistry B, 123, 983–993 (2019).

Histone tails are integral structural and functional components of the eukaryotic nucleosome. These tails, rich in positively charged amino acid residues, interact with the DNA to stabilize the nucleosomal structure. However, capturing the biochemical effects of posttranslational modifications (PTMs) on histone tails in molecular detail using X-ray or NMR techniques remains challenging due to their intrinsically disordered structure.
Using multiscale modeling techniques, we are quantifying the conformational and structural changes that occur when histone tails undergo methylation, phosphorylation, or acetylation.
Key Publications:
Under Review.
Proteins are workhorses of biology perform uncountable functions to sustain life. Composed of 20 key amino acid residues, the proteins adopt secondary, tertiary, and quaternary structures based on their primary sequence, physiological conditions, and solvent.
We are interested in predicting protein-protein assembly as a result .
